Quantifiler Human Dna Quantification Kits User's Manual
4343895 Used With: Quantifiler™ Human DNA Quantification Kits Users Manual, Quantifiler™ Y Human Male DNA Quantification Kit 2.00% $1,082.72 $1,0790 Quantifiler™ Human DNA Quantification Kits User's Manual 2.00% $217.15 $2906 Quantifiler™ Y Human Male DNA Quantification Kit 2.00% $1,082.72 $1,0 HID ACCESSORIES.
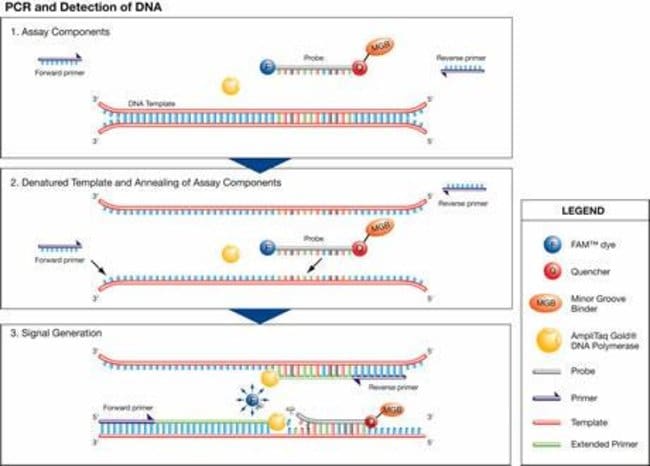
The quantity of DNA was determined by Quantifiler Duo, Quantifiler Human, and Quantifiler Y Human Male DNA Quantification Kits (Applied Biosystems). Real-Time PCR Amplification Real-time PCR amplification reactions contained 10.5 lL of Primer-Probe Mix, 12.5 lL of Master Mix, and 2.0 lL of DNA sample. Quantifiler Human DNA Quantification Kit User's Manual. Applied Biosystems, Foster City, California, 2003. Budowle B, Moretti TR, Keys KM, Koons BW, Smerick JB. Validation studies of the CTT STR multiplex system. J Forensic Sci 1997; 42(4):701-707. Buse EL, Putinier JC, Hong MM, Yap AE, Hartmann JM. Performance evaluation of two multiplexes. Quantifiler ® Duo DNA Quantification Kit: A Guiding Tool for Short Tandem Repeat Genotyping of Forensic Samples. Quantifiler ® Duo DNA quantification kits User's Manual, PN 4391294 Rev.B. The Quantifiler® Kits User’s Manual provides information about and instructions for using the Quantifiler® Human DNA Quantification Kit and the Quantifiler® Y Human Male DNA Quantification Kit. Text Conventions This guide uses the following conventions:. Bold indicates user action. The Quantifiler Duo DNA Quantification Kit enables forensic laboratories to simultaneously obtain a quantitative and qualitative assessment of total human and human male DNA in a single, highly sensitive real-time PCR reaction. This guides selection of the optimal STR chemistry (autosomal, Y-STR.
Abstract
Analysis of the length polymorphisms of short tandem repeats (STR) loci in the human genome has become a standard approach for comparative genotyping in many areas including disease research and diagnostics, parentage assessment, investigations of human diversity, and forensic science. The purpose of this study is to optimize the DNA concentration in ng/10μL for amplification of DNA markers. AmpFlSTR Identifiler Kit is used to amplify STR markers and capillary electrophoresis is used to analyze DNA profile of human the genome. Two sets of samples with following DNA concentration: 100 pg – 6 ng/25 μL were used for this study. There was no DNA profile detected in samples with concentrations 100 pg - 300 pg/25 μL (pictogram), while in some cases partial DNA profile was yielded. On the other hand samples with 0.4 ng – 4 ng/25 μL, yielded a full DNA profile. We were not able to obtain any profile using concentrations over 4 ng/25 μL. Improvements in detection limits/sensitivity at upper and lower DNA concentrations are of potential benefits to amplify STR of Human Genomic in order to obtain a full DNA profile. The optimal DNA concentrations which produced reliable and balanced peaks, no off scale peaks and full DNA profile for all loci were at range 0.4 ng – 3 ng/25 μL.
Quantifiler Human Dna Quantification Kit
INTRODUCTION
Eukaryotic chromosomal DNA, polymorphic short tandem repeat (STR) loci are key tools for: rapid gene discovery, disease locus mapping and carrier diagnosis of disease states, linkage analyses, agricultural genetics, parentage assessment, and population diversity studies. The ability to detect genetic differences between individuals increases when DNA typing information at multiple polymorphic STR loci is combined [1, ]. Clinical diagnostic laboratories usually perform analyses of biological samples that have been collected and stored in ideal conditions, making sample quality and quantity rarely an issue. Those ideal conditions are usually not met in the DNA typing analysis of forensic biological evidence, as there is no control over the amount of biological material left at a crime scene or its level of degradation or contamination due to exposure to various environmental insults [3, ]. Forensic biological samples may present challenges for DNA typing analysis, even with the utmost care throughout crime scene evidence recovery and storage. Due to potential exposure to a virtually unlimited number of uncontrolled variables, forensic casework specimens may result in particularly challenging polymerase chain reaction (PCR) templates. Optimized concentration of DNA and reagents is very important on PCR amplification of different combinations of 15 polymorphic tetranucleotide STR loci in a single reaction tube. This study has done to check the upper and lower limits of DNA concentration used in amplification step to obtain full DNA profiles [5].
MATERIAL AND METHODS
Samples
Our study was performed using human blood as biological material. Before analysis the blood sample has dried on filter paper. The size of the samples used for extraction were 3×3 mm2 and Chelex-100 resin is used as extraction method [6].
Procedures
For quantification purposes Absolute Quantitation method is used, real-time quantitative PCR assay with a fluorogenic TaqMan® probe targeting the human telomerase reverse transcriptase gene (hTERT) and supported by Quantifiler® Human DNA Quantification Kit on Instrument ABI PRISM® 7000 Sequence Detection System (Applied Bioystems-AB, P/N 4330087 with SDS Software v1.0) [7, 8]. For amplification purposes we introduced AmpFlSTR® Identifiler® PCR Amplification Kit (AB P/N 4322288) [5]. The following concentrations (ng/10 μL) of template DNA were amplified in a total reaction volume of 25 μl: 0.1, 0.2, 0.3 up to 6 ng. Samples were amplified in 0.2-ml microAmp reaction tubes with caps (AB P/N N801-0540) using the following conditions (recommended by the manufacturer, Applied Biosystems): 95°C for 11 min, 28 cycles of 94°C for 1 min, 59°C for 1 min and 72°C for 1 min, 60°C extension for 60 min and 2-8°C until samples were analyzed further. The amplification was carried out on GeneAmp PCR System 9700 (AB P/N N805-0001) [5], [1]. The amplified DNA markers were: D8S1179, D21S11, D7S820, CSF1P0, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, Amelogenin, D5S818 and FGA [5], [1]. Finally detection of amplified samples was done using 1.5 μl of amplified DNA product that was combined with aliquot 25 μL of the Hi-Di™ Formamide (AB P/N 4311320) mixed with GeneScan™ 500 LIZ™ Size Standard (AB P/N 4322682). Capillary electrophoresis was carried out using an automated DNA sequencer ABI PRISM® 310 Genetic Analyzer, AB P/N 310-00-200/240-W. The raw data were analyzed using GeneMapper® ID Software v3.2. The minimum peak height threshold was set at 50 RFU [9, 10].
Quantifiler Human Dna Quantification Kits User’s Manual
RESULTS
Samples with DNA concentration at 0.1 and 0.2 ng/25 μL yielded no DNA profile (Figure 1). No DNA profile was obtained from samples with 0.3 ng/25μL, however in this case some loci were included (Figure 2). Baseline had noisy and allelic imbalance as well. Samples with DNA concentration from 0.4 - 3 ng/25μL yielded full DNA profiles (Figure 3). There were no any allelic imbalance, stutter or any other artifacts except for the sample concentration at 3 ng/25μL. Samples with concentrations 2.7 and 3 ng/25μL exhibit decrease of Relative Fluorescence Units (RFU) starting from shorter loci toward longer ones for all panels. The loci with lower RFU were D7S8210, CSF1PO, D2S1338, D18S51 and FGA [11].
Presents electropherogram of sample with No DNA Profile analyzed with AmpFlSTR Identifiler Kit. There is no any allele obtained in total of 16 human genome loci analyzed.
Presents electropherogram of samples with Partial DNA Profile analyzed with AmpFlSTR Identifiler Kit. There are three alleles obtained in total of 16 human genome loci analyzed.
Presents electropherogram of samples with Full DNA Profile analyzed with AmpFlSTR Identifier Kit. Samsung s8 cellular data. There are all alleles obtained in total of 16 human genome loci analyzed.
Ps4 cuh 1001a motherboard. View and Download Sony PlayStation 4 user manual online. PlayStation 4 Game Console pdf manual download. Game Console Sony PlayStation 4 CUH-1001A Quick Start Manual. Video game console (2 pages). When different options on the menu pop up, choose the option called “turn off the PS4.” This will allow you to turn off the device, which. View and Download Sony PlayStation 4 CUH-1001A quick start manual online. Video Game Console. PlayStation 4 CUH-1001A Game Console pdf manual download. Also for: Ps4 cuh-1001a. Getting started Turn on the TV, and then set the TV input to HDMI. Touch the (power) button of the PS4™ system to turn the system on. The power indicator blinks in blue, and then lights up in white. Connect the DUALSHOCK ®4 wireless controller to the system using the USB cable, and then press the (PS) button. Find out the online pdf manual for setting up your PlayStation 4 game console. Also check out the manuals for all the compatible peripherals, safety and support guide, troubleshooting, specifications, and other information.
DNA concentration between 3 - 4 ng/25 μL yielded full DNA profiles but there were allelic imbalance peaks and significant difference on RFU in heterozygote peak height ratios between the shorter and longer loci (Figure 4).
Presents electropherogram of samples with Full DNA Profile analyzed with AmpFlSTR Identifier Kit. There are all alleles obtained in total of 16 human genome loci analyzed. At some alleles there are some imbalance peaks.
For shorter loci RFU was over 1500 to 3500, whereas for longer ones were under 500. Despite these results, DNA profile obtained from above samples can be used officially as DNA Profile at System of Justice. No DNA profile was yielded in samples over 4 ng/25μL (Figure 1). Some loci were obtained, but just few of them (below 5) so they did not represent any relevance in DNA profile. To be valid the DNA profile must have at least 10 loci obtained. Those profiles with number of loci below 10 are unavailable to be used for any purpose [1], [9]. The referent samples yielded a full DNA profile. The best DNA profile was obtained from samples with DNA concentration at 0.5 – 1.8 ng/25 μL. Reliable DNA profiles were obtained from samples with 0.4, 0.5 and 2 – 4 ng/25 μL as well (Table 1).
TABLE 1
Represents DNA profile yielded on different DNA concentrations
DISCUSSION
A variety of commercial Kits for DNA amplification are now available and they routinely are used in forensic and diagnostic laboratories worldwide. Although generally reliable and convenient, many of these Kits offer only a relatively limited dynamic range for upper and lower limits, so that in many cases samples from the crime scene, reference samples and clinical samples frequently have to be diluted or concentrated and retested in order to avoid exceedi ng the upper or lower limits. At the same time analyzes is performed by using real-time PCR which is time and money consuming. The advantage of the current study is the fact that we found that upper and lower limits beyond of what is actually suggested by companies. This method increases the opportunity to amplify DNA directly from extraction step into amplification one without any trouble toward result interpretation. DNA yielded profile from an amount up to 4 ng/25 μL (which is a double amount suggested by protocols), crucial due to the fact that is easy to determine that approximate amount in our sample size during the extraction step. This decreases a workflow of our analysis which in this case will consist of extraction, amplification and detection without quantification step. This workflow would not have any interference in terms of detecting sensitivity of DNA. In addition, this method can serve as a great tool to help laboratories in obtaining reliable DNA profiles by saving time and money. Leclair et al. have studied lower and upper limits for DNA concentrations and total volume used for DNA amplification. In this study the best results were yielded using 0.5 ng of DNA in a 10-μL PCR reaction volume which appeared to produce an identical profile to 2 ng of the same DNA in a 40-μL reaction volume []. Company Applied Biosystems which has performed a validation of the AmpFlSTR Identifiler Kit, suggested using an amount of DNA concentration in the range of 0.5 - 2 ng/25 μL [5].
CONCLUSIONS
AmpFlSTR Identifiler, the PCR Amplification Kit used to amplify STR markers of analyzed samples exhibited to be very stable. Primers of this Kit were sensitive and very specific for DNA markers as mentioned above. Even if AmpFlSTR Identifiler PCR Amplification Kit – User manual suggests using DNA concentration 0.5 – 1.2 ng/25 μL (lower and upper limits) [5], and National Institute of Standards and Technology (NIST) suggests 0.5 – 2 ng/25 μ L as the best range of STR Kits [1]. We have obtained full DNA profile with a lower concentration of 0.4 ng/25 μL up to 4 ng/25 μL for the upper one. Other reagents of this Kit: AmpFlSTR® PCR Reaction Mix, AmpliTaq Gold® DNA Polymerase and AmpFlSTR® Identifiler™ Allelic Ladder used on 15μL as a master mix shown to be enough for amplification of samples with concentration 4 ng/25μL without any problem for inhibition by the template. Performing the step of DNA quantification on RT-PCR is time consuming and cost effective. We found that the best and reliable results are yielded beyond the limits suggested by different companies. We obtained reliable DNA profile from concentration up to 4 ng per sample analyzed. Using those results, the laboratories can make validation of methods in different applications such as: molecular biology, genetics, biomedicine etc to analyze samples directly from extraction to amplification and by skipping quantification step in RT-PCR. By applying such approach it could be possible to save time and money.
ACKNOWLEDGMENT
We would like to thank you Nancy Whitney Peterson (Forensic Biology/DNA Consultants, LLC, USA) for constructive criticism and suggestions in the preparation of the manuscript.
DECLARATION OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES